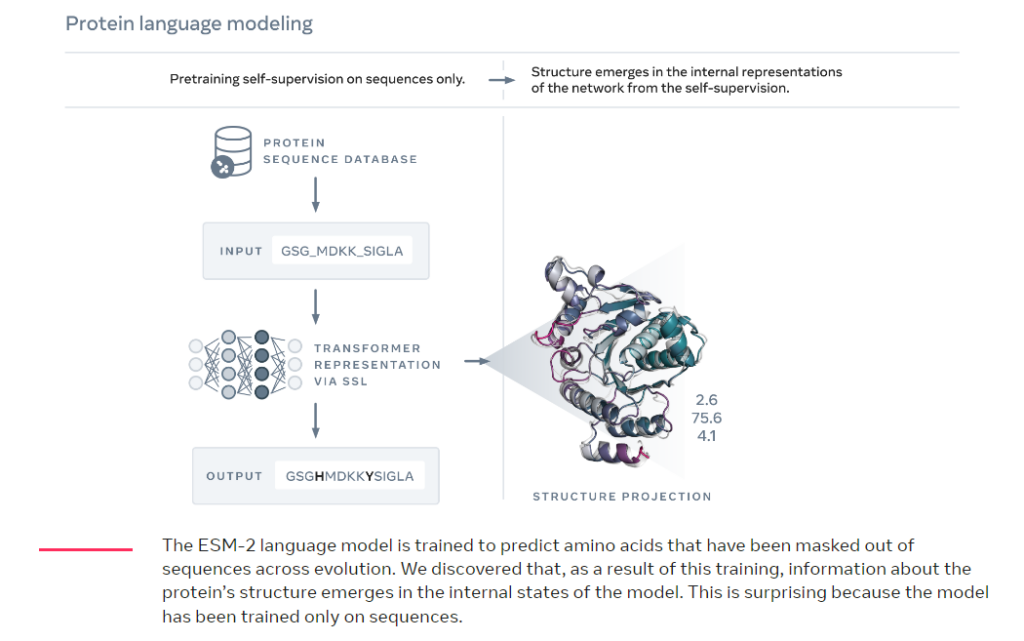
ESMFold (ESM = Evolutionary Scale Modeling) [paper] uses a large language model that allows to accelerate folding (i.e. predicting the 3D structure of a protein from the DNA sequence [that encodes the amino acid sequence]) by up to 60 times (compared to state-of-the-art techniques like AlphaFold). This improvement has the potential to accelerate work in medicine, green chemistry, environmental applications, and renewable energy.
In addition, Meta AI made a new database of 600 million metagenomic protein structures (proteins which are found in microbes in the soil, deep in the ocean, and even in our guts and on our skin) available to the scientific community via the ESM Metagenomic Atlas.
ESMFold and related models like ESM-2 are published together with the API on GitHub and HuggingFace.

Leave a Reply