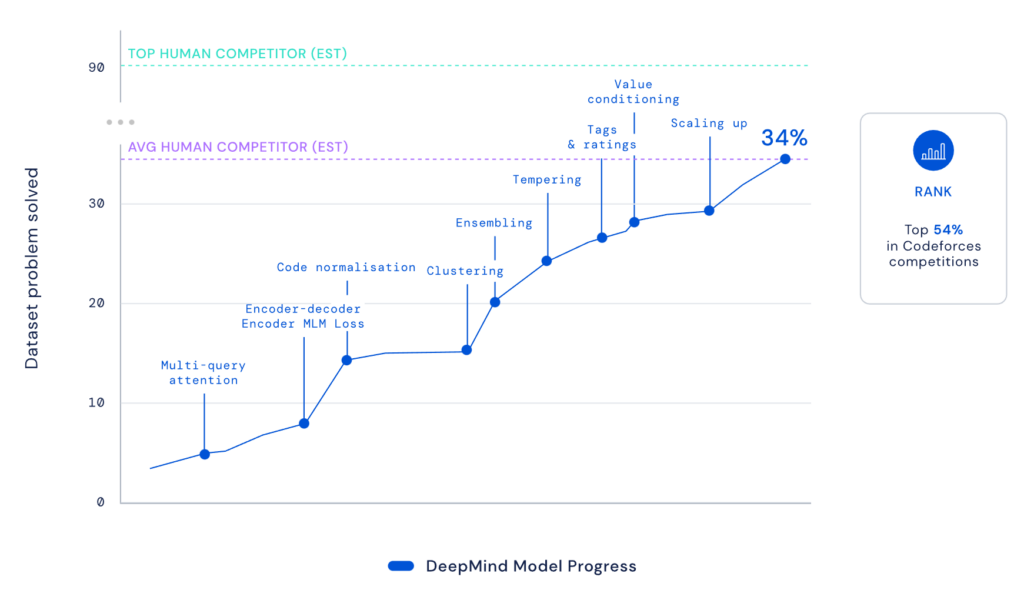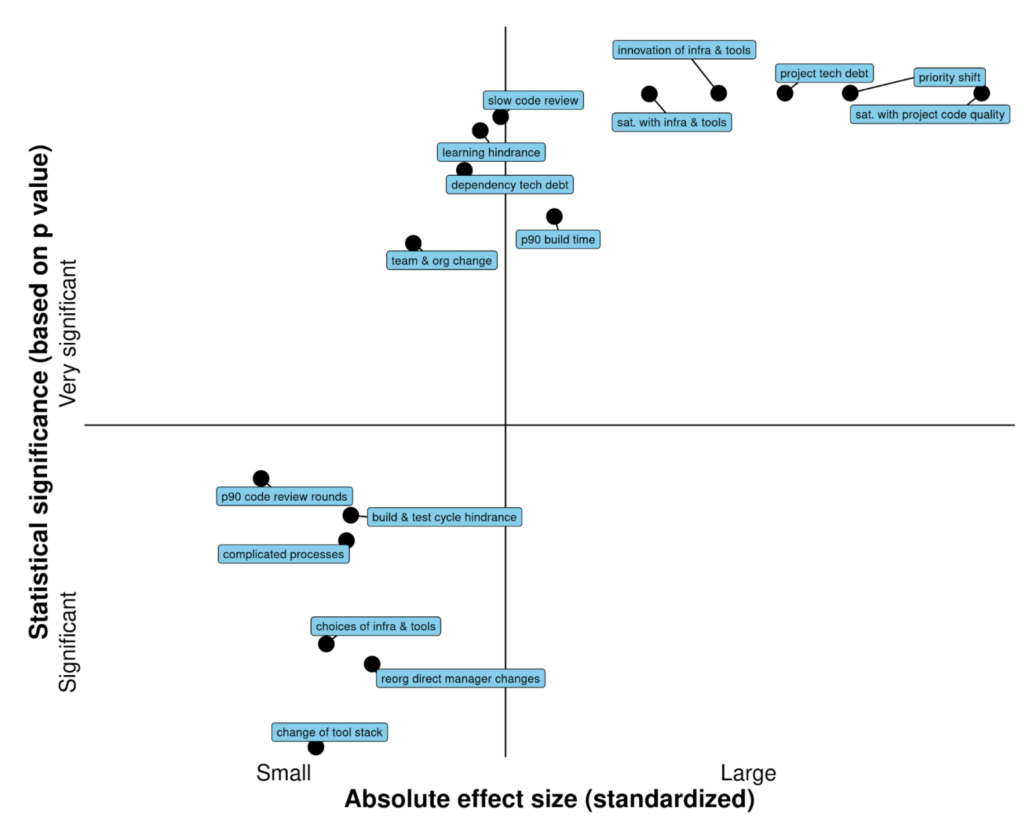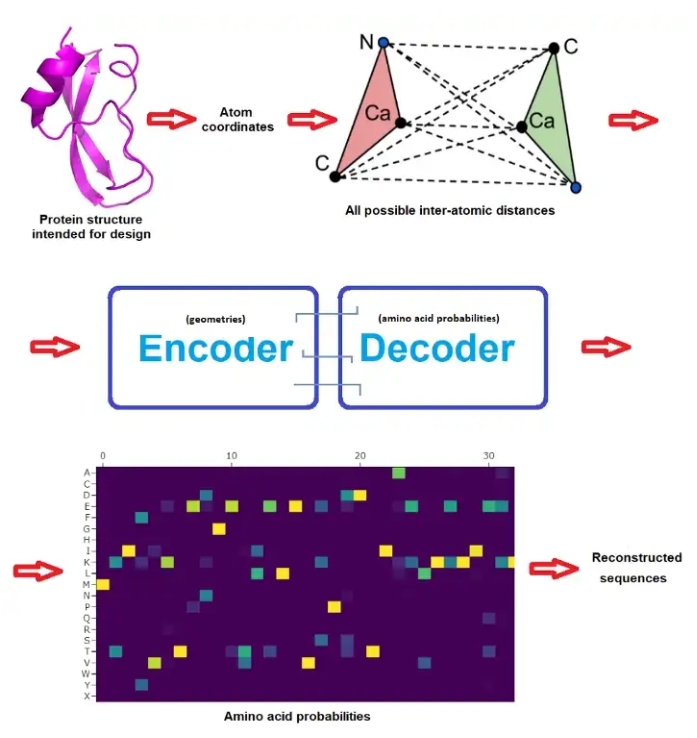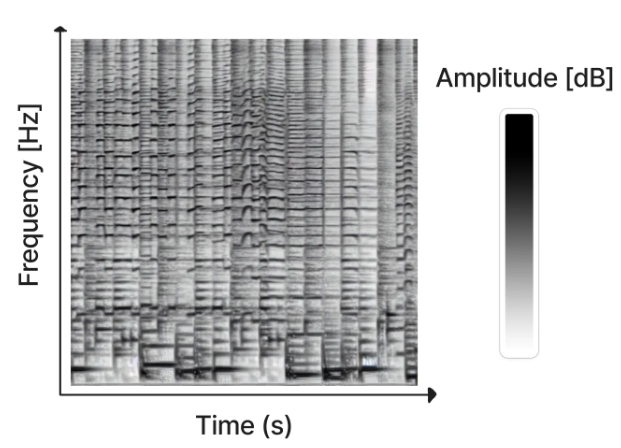
By using the stable diffusion model v1.5 without any modifications, just fine-tuned on images of spectrograms paired with text, the software RIFFUSION (RIFF + diffusion) generates incredibly interesting music from text input. By interpolating in latent space it is possible to transition from one text prompt to the next. You can try out the model here.
The authors provide source code on GitHub for an interactive web app and an inference server. A model checkpoint is available on Hugging Face.
There is a nice video about RIFFUSION by Alan Thompson on youtube.
Even more shocking than using diffusion on spectrograms and getting great results may be a paper by Google Research published on Dec 15, 2022. They use text as an image and train their model with contrastive loss alone, thus calling their model CLIP-Pixels Only (CLIPPO). It’s a joint model for processing images and text with a single ViT (Vison Transformer) approach and astonishing performance.